The Summer Undergraduate Research Creativity Fellowships provide undergraduates with an opportunity to study a wide variety of disciplines through self-directed work under a faculty mentor. The Student Sustainability Council collaborates with the Office of Undergraduate Research in order to promote projects that will make a significant contribution to the student’s academic growth while simultaneously contributing to the sustainability-focused research initiatives at the University of Kentucky and the community beyond. This collaboration aids in making the pursuit of sustainability goals an integral part of the UK student experience, which is one of UK’s sustainability guiding principles.
Katie Christensen
Dr. Kendall Corbin
The majority (∼75%) of today’s antibiotics are derived from natural products, including compounds isolated from fungus and bacterium (1). Unfortunately, due to a range of factors, resistance has been observed for nearly all antibiotics that have been developed, leading to a global antibiotic resistance crisis (2). One means in which to help combat this crisis is to identify novel antibiotic producing microorganisms.
Soil has been found to be a major source of microorganisms that produce antibiotics, likely due to its microbial diversity and richness. However, lesser studied ecosystems like plant biomass, have largely been ignored, resulting in a potentially untapped source of novel antibiotic producing microorganisms. This theory is further supported by recent findings, which showed that plant leaf surfaces are abundant in Streptomycetaceae (3), the family of bacteria responsible for producing over 75% of clinically useful antibiotics.
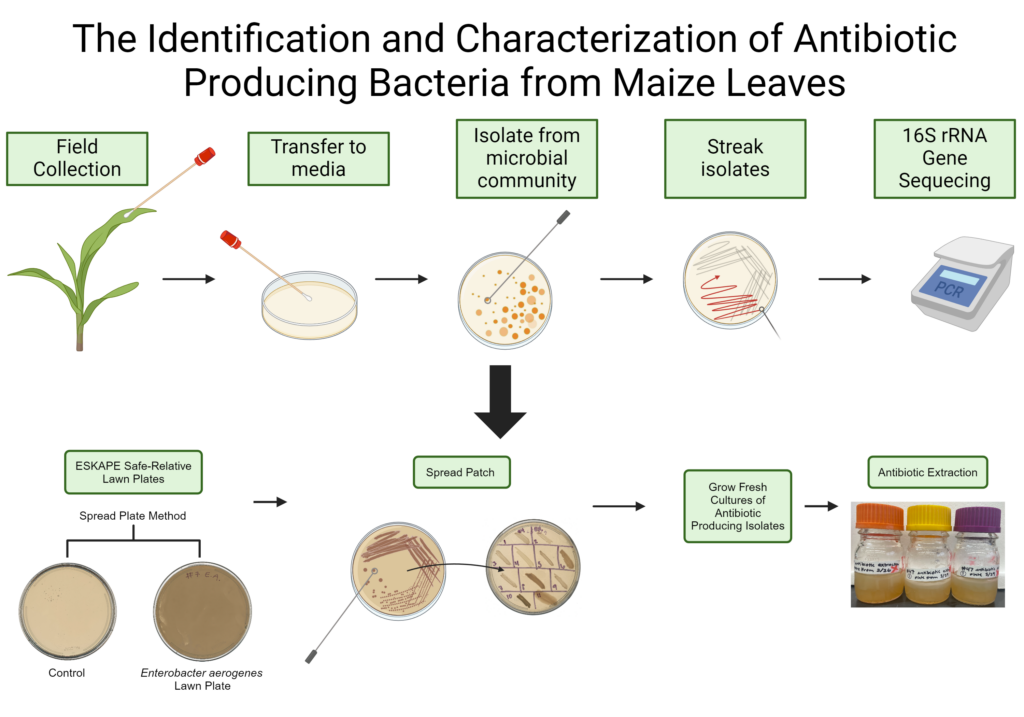
In this study, bacteria from the top and bottom of 5-week-old corn leaves were collected. The swabs used for collection were diluted and plated on 4 types of media. The 4 media types of media used were Tryptic Soy Agar (TSA), Luria Broth (LB), All Culture (AC), and Potato Dextrose Agar (PDA). The streak-plate method was used to isolate pure cultures of bacteria from the plates. In total 225 bacterial isolates were collected for this study.
For my project this summer, I will 1. determine the identity of each isolate and 2. investigate the ability of unique isolates to produce compounds of interest (e.g. antibiotics). The identity of the isolates will be determined by amplifying the 16S ribosomal gene using polymerase chain reaction (PCR). The PCR technique used in this project will be the colony PCR bead method. This method allows the bacterial isolate DNA to be amplified without DNA extraction. After amplification, the PCR product will be sequenced using Sanger sequencing and a taxonomical assessment made using bioinformatic tools. Selected species of interest will be tested against Enterococcus raffinosus, Staphylococcus Epidermidis, Escherichia coli, Acinetobacter baylyi, Pseudomonas putida, and Enterobacter aerogenes to test for antibiotic properties. These 6 bacterial species are the safe relatives of the pathogens Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species (ESKAPE). I will use chemical separation techniques to extract the compounds of interest and determine if antibiotic activity is maintained using different assay techniques.
If successful, this project will identify unique species of bacteria from corn leaves that exhibit antibacterial properties. Findings generated from this study will help bring awareness to the importance of studying microbially diverse ecosystems, like plant biomass, for antibiotic discovery. ESKAPE pathogens are multi-drug resistant and highly virulent, causing havoc in a world that has slowed antibiotic production. Overall, this project has the potential to help combat the growing global crisis of multi-drug resistant ESKAPE pathogens.
(1) https://www.pnas.org/doi/full/10.1073/pnas.1618221114
(2) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4378521/
(3) https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-021-01118-6
(4) Hernandez, et al., (2022). “A Research Guide to Studensourcing Antibiotic Discovery.” XanEdu Publishing Inc.